
Building a python library with the Pre-Trained models has been one of my secondary objectives for GSoC 2020, and it was about time that I got started with it. Devolearn is a python library that can be used to collect useful metadata from videos and images of C. elegans embryos with deep learning.
Here’s a quick example with which one can get started with DevoLearn:
Installation
pip install devolearn
Segmenting the C. elegans embryo
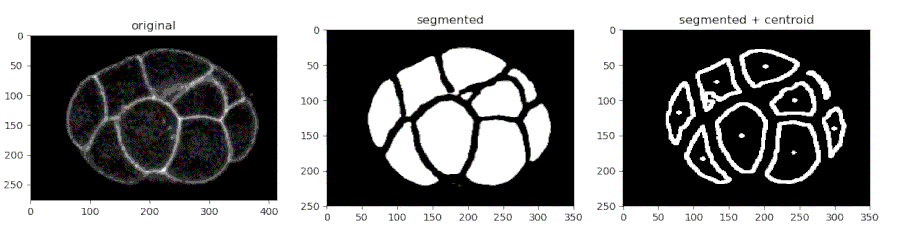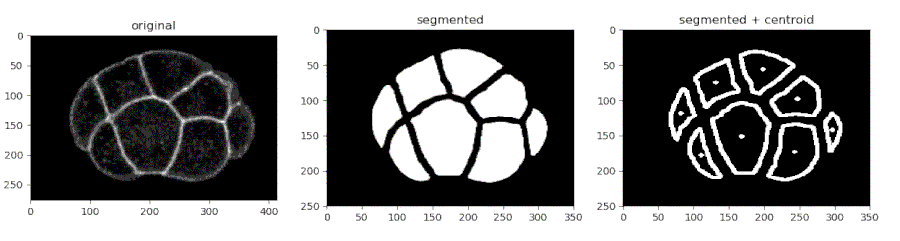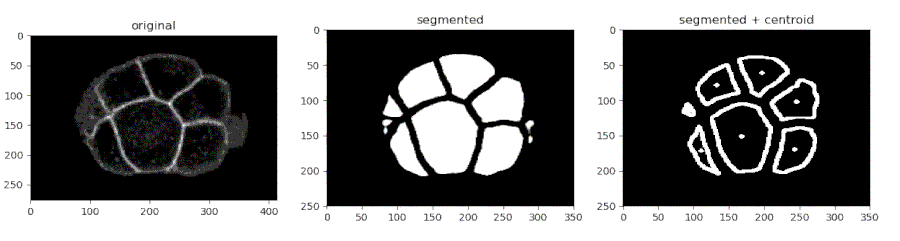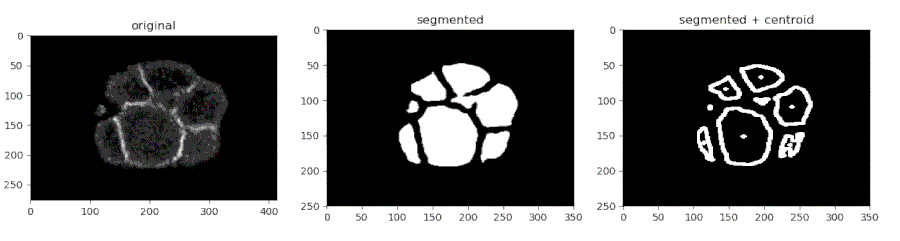
- Importing the model
from devolearn import embryo_segmentor
segmentor = embryo_segmentor()
- Running the model on an image and viewing the prediction
seg_pred = segmentor.predict(image_path = "sample_data/images/seg_sample.jpg")
plt.imshow(seg_pred)
plt.show()
- Running the model on a video and saving the predictions into a folder
filenames = segmentor.predict_from_video(video_path = "sample_data/videos/seg_sample.mov", centroid_mode = False, save_folder = "preds")
I won’t be including all of the examples here, because it’s already there on the README.md of the devolearn repo.
As you might be able to see, the primary focus of devolearn was ease of use, the user could be a complete beginner in python and still be able to use the deep learning models in devolearn for his/her research. All that he has to do is to know the filename of his images/videos which he wants to work on.
One can use the embryo_segmentor() to extract the centroids of the cells from a video and save the centroid co-ordinates into a CSV file which might look like this.
Devolearn is open to any form of contributions/improvements from the open source community. The best place to start for someone who wants to contribute is the contributing.md file.
I’ll be writing more on how devolearn works under the hood in the next few weeks.