The bridge works now
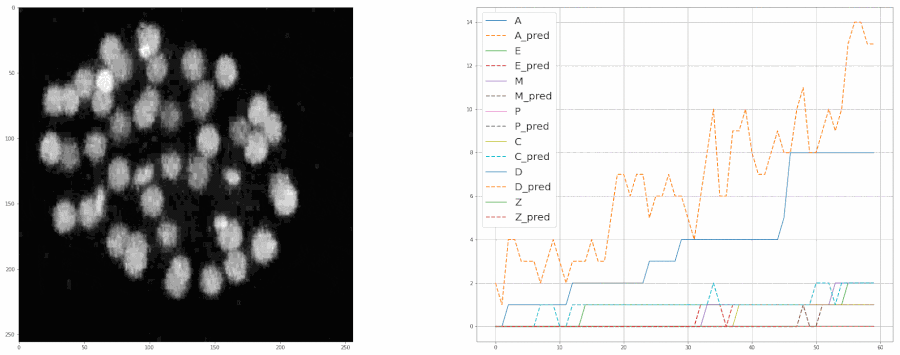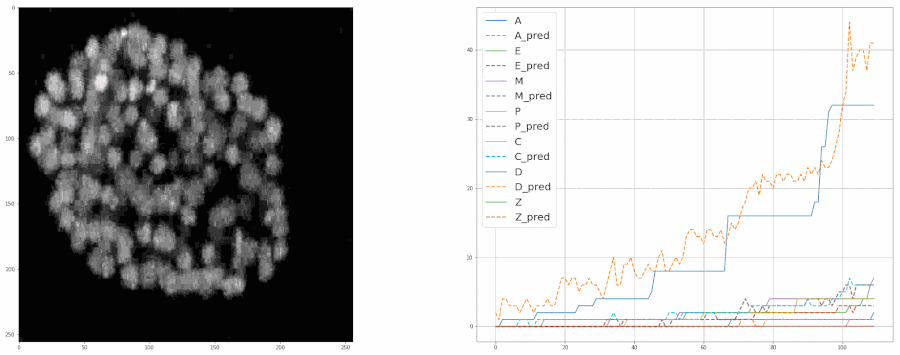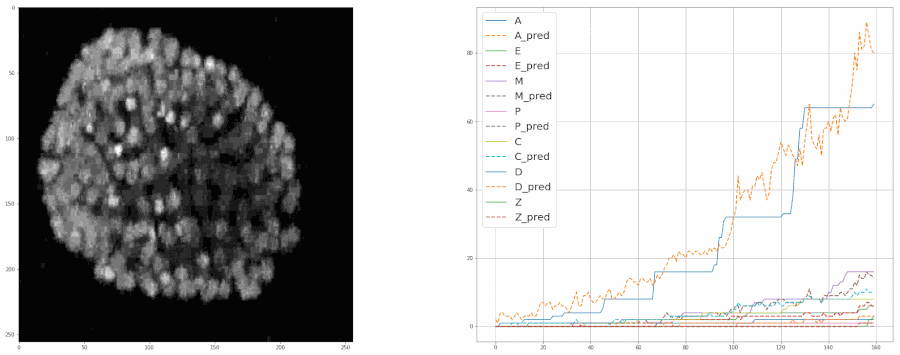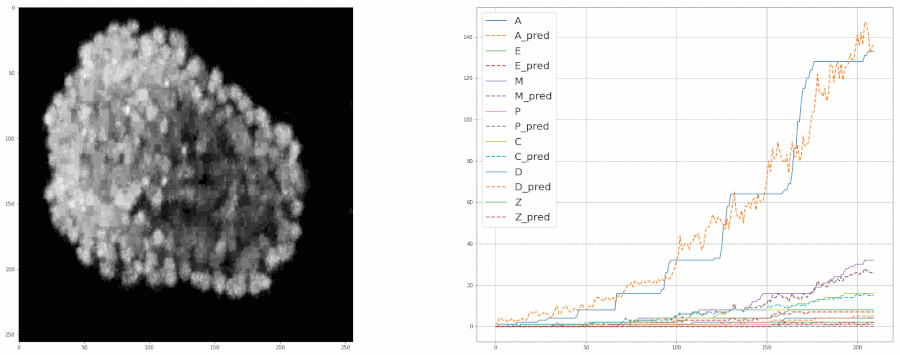
So, after cleaning up the data last week, I started training the model again from scratch. This time I included all the bells and whistles like ReduceLROnPlateau and image augmentation with albumentations. Check out this notebook for the code.
All the values of the populations were yet again normalised to values between -1 and 1. The ResNet18 was customized to a smaller last layer (7 in our case, because 7 lineages of cells).
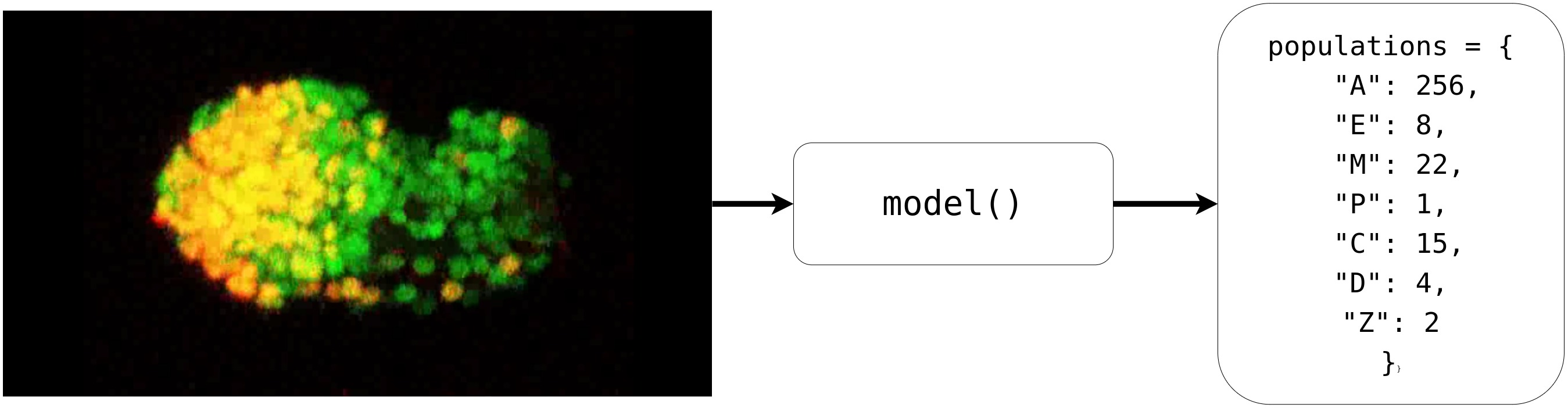
And after training the model for 8 epochs, the model was able to estimate the populations pretty well. So much so that it was almost at par with the accuracy of the real data.
We fed a whole timelapse of the embryogenesis of the C. elegans embryo into the trained model and plotted its predictions, and compared it with the real values as shown:
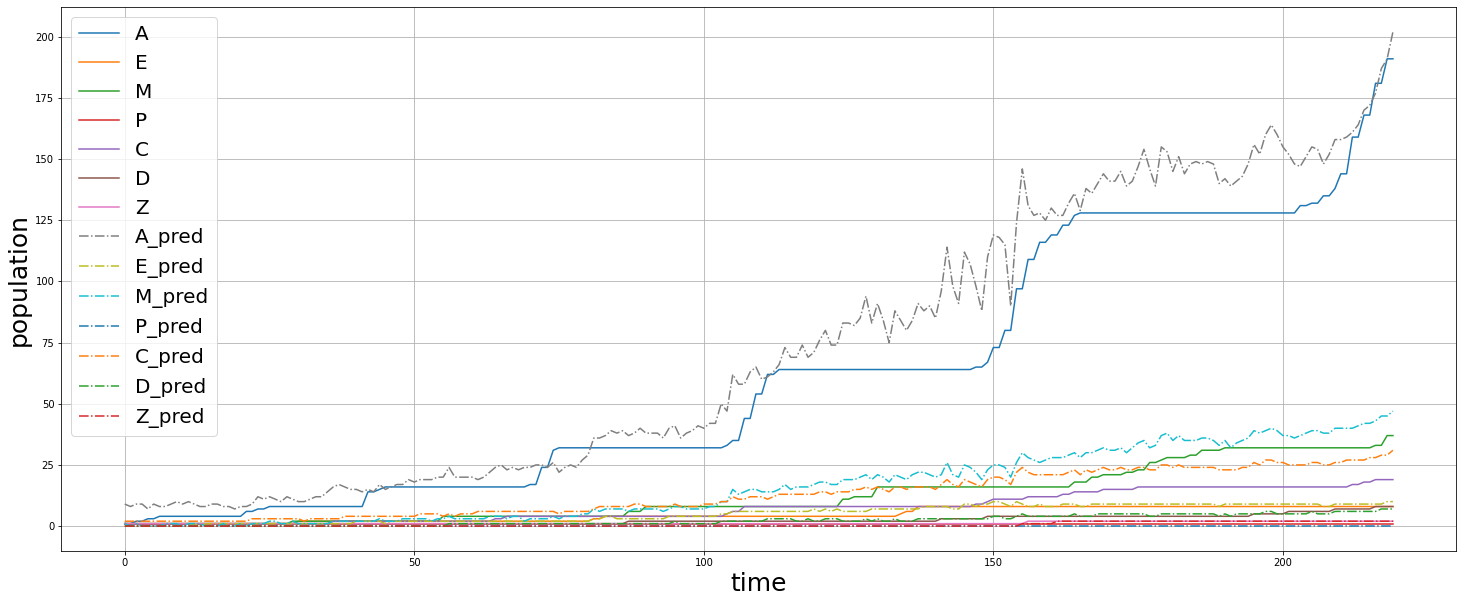
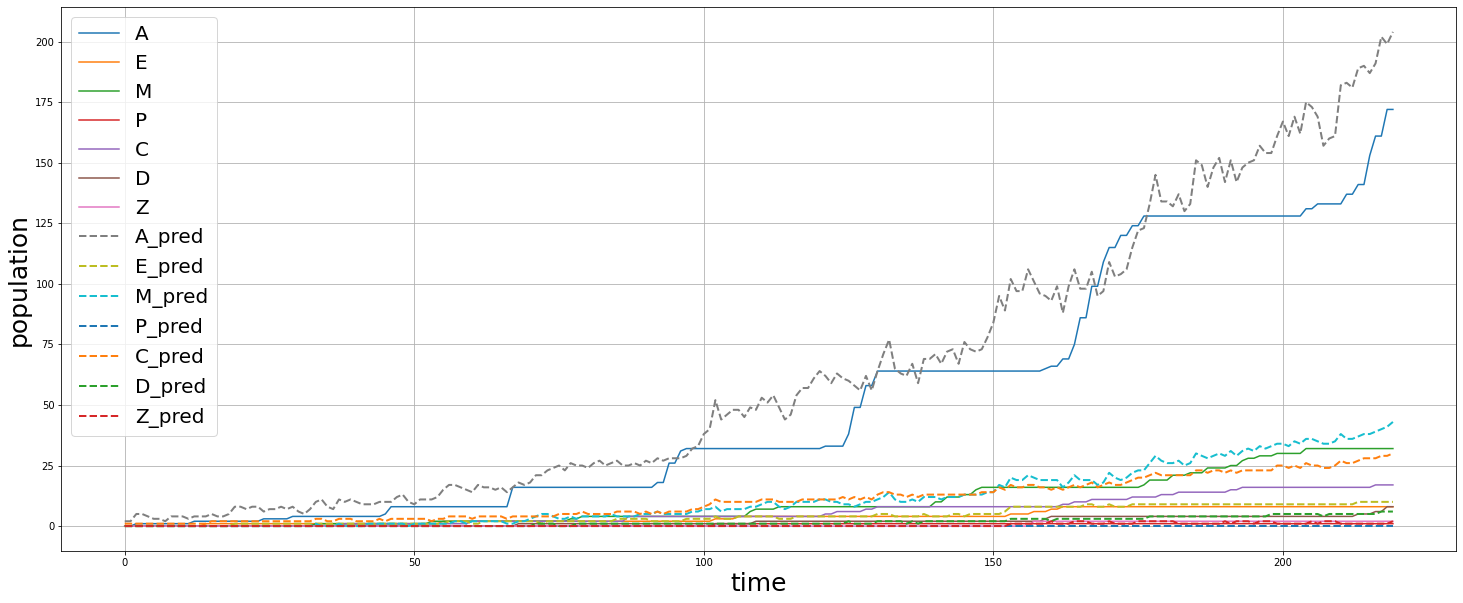
Py_elegans
Apart from all of this, I also started working on a python library, which would make these deep learning models I’ve been training more accessible to the commmunity through a high level framework. For now it’s temporarily named “py_elegans”.
In this framework, loading up a pretrained model would be as easy as
from pyelegans import lineage_population_model
and predictions can be made using:
pred = model.predict(image_path = "sample.png")
More updates on this next week.