The bridge that we’ll have to build is between the videos and the annotations in the EPIC dataset, and for that we’ll need to train a deep learning model that connects these two. It turned out that it wasn’t as straighforward as throwing a bunch of tensors into a model.
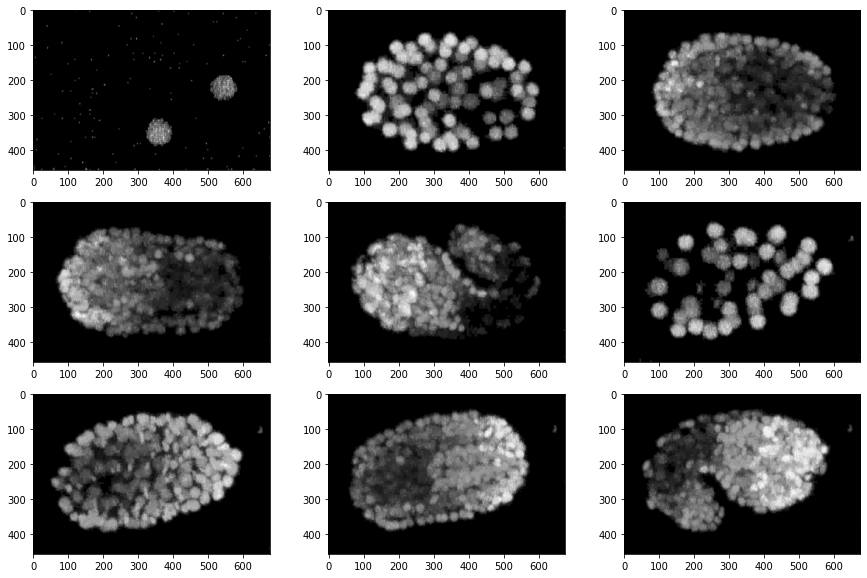
From the frames like the ones shown above, we’ll have to find the population of various lineages of cells within the embryo.
The C. elegans embryo contains cells from various lineages which move on to become various body parts during adulthood, and the total population of each lineage depends primarily on time, but there are a few other biological factors as well. The plot below shows how the populations of each lineage varied with time for a single embryo.
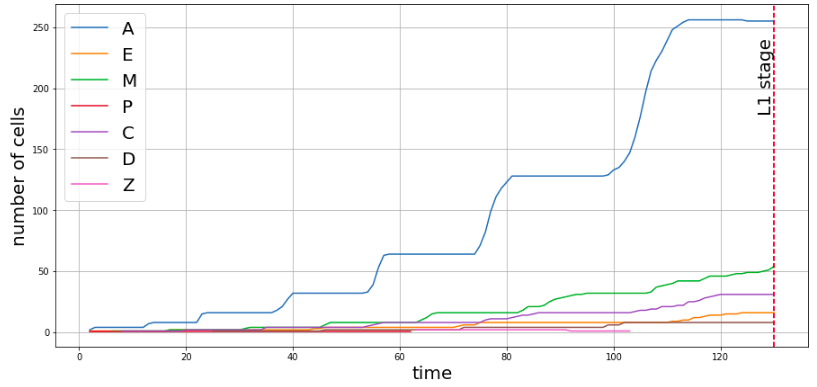
The first task was to format the data in a way that can be used to train a model to gain inferences. This mostly involved algorithms that determined the population of each lineage with time from the raw metadata.
The second problem was with mapping the video with the annotations. The issue was that there were about 130 or so time instances per video, but there were around 40 frames. So I had to write another algorithm that distrubutes these 240 frames on the 130 time instances.
So now all that was left was to train a deep learning model which would map the frames to the populations. The prediction for each frame of the model is a 1 dimensional tensor which contains the populations of the lineages ['A', 'E', 'M', 'P', 'C', 'D', 'Z'] in that order.
When trained on only 2 videos and about 480 frames, these are the results that were obtained:
