Ten weeks of proposal writing and 3 personal (but very beloved) deep learning projects later, I was selected for the only project I worked towards for GSoC, which was Pre-trained models for developmental neuroscience under INCF. And things have been better than ever since then.
The first week was mostly me getting to know more about the OpenWorm community and what are they striving to do. I got to meet Dr. Bradley Alicea, Vinay Varma and Ujjwal Singh. They gave me a warm welcome into the community.
I started setting up the environment right after the results were out, and cleaned up one of my older repos which would act as a quick reference through the span of this project.
The first weekly meeting with OpenWorm gave me a much better idea of the goals of this project.
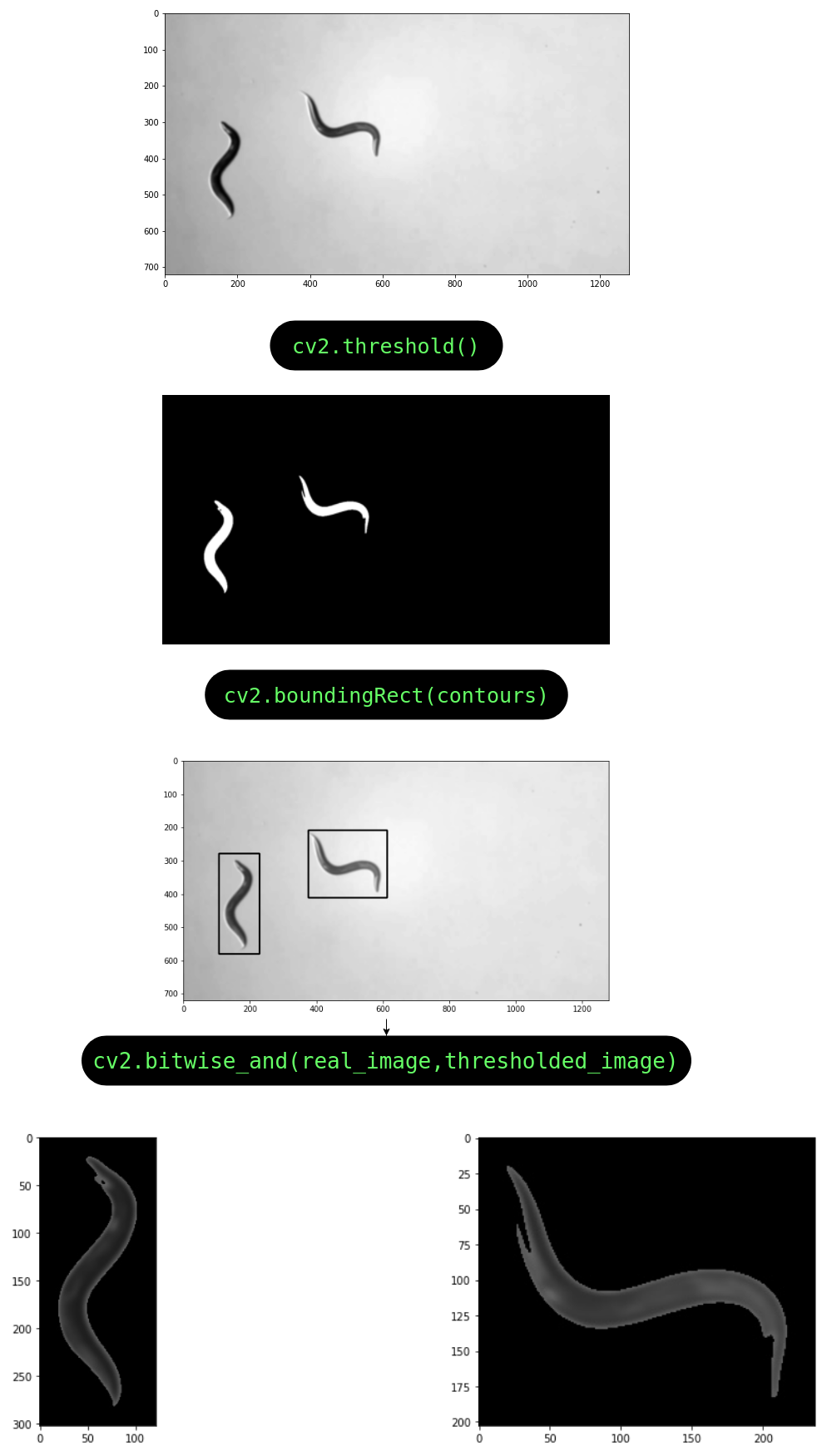
The key to a good deep learning model is good data. If the data is clean, half the battle is won. This led to the the first notebook which builds a pipeline for tracking and segmenting the worms directly from video files. The pipeline works as follows:
-
The video file is first trimmed to shorten the clip to extract only a short subclip (this is optional).
-
All the frames from the subclip are extracted from the video using ffmpeg and saved into a folder in a numerical sequence of filenames.
-
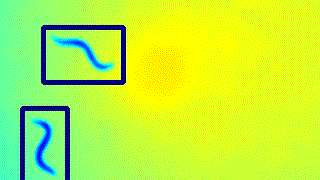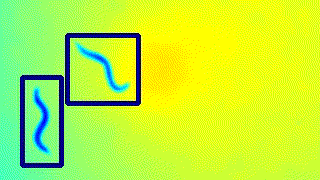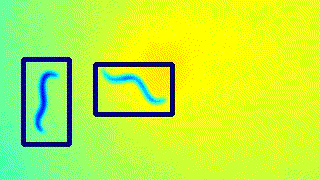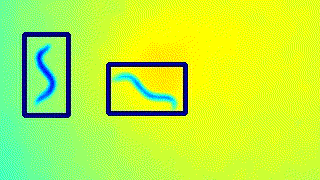
The locations of the worms are determined by drawing padded rectangles around the contours of the thresholded image.

- The original image is then cropped and segmented to remove all background except the worm(s) in the image.

Here’s a GIF to visualise the tracking on the live feed.
Apart from all of this, I also read quite a few papers on the C.elegans worm. This was an effort towards getting more familiar with the underlying biological concepts surrounding the worm.